forum@abinit.org
Subject: The ABINIT Users Mailing List ( CLOSED )
List archive
- From: Chol-Jun Yu <yucj@ghi.rwth-aachen.de>
- To: forum@abinit.org
- Subject: Re: [abinit-forum] B2 NiAl
- Date: Wed, 20 Feb 2008 18:33:21 +0100
I have attached some related data which include the strained unit cells and reference how to calculate the elastic constants.
Hope helpful.
Chol-Jun
Stephen.Hocker@mpa.uni-stuttgart.de wrote:
Dear Abinit users,
I'm trying to calculate the elastic constants of B2 NiAl using the serial
version of Abinit 5.4.4, unfortunately I didn't obtain any results yet. Some
values for the parameters in my infile must be wrong, but I have no idea
which ones I should change.
The calculation starting from an input file like the one for Al which is given in the elastic tutorial stopped with the message:
kpgsph : BUG -
The variables ikg, mkmem, and mpw must satisfy ikg<=(mkmem-1)*mpw,
while the arguments of the routine are
ikg = 3060, mkmem = 20, mpw = 159
Probable cause: Known error in invars1 for parallel spin-polarized case.
Temporary solution: Change the number of parallel processes.
Action : contact ABINIT group.
Therefore I decided to determine the elastic constants by calculation of the
energies of strained samples. In these calculations the total energy should increase parabolically with increasing strain. But it doesn't. Depending on the parameters it usually increases somehow, but the curve is never a parabola. In some cases it even decreases a little. These are the parameters I already tried:
ecut (280...480 eV)
ngkpt (2 2 2...10 10 10)
nband/fband 65/0.0d0 occopt 3...4
tsmear 0.03...0.07
I tried with different pseudopotentials (Troullier-Martins) as well, namely
13al.pspnc, 28ni.pspnc / 13-Al.LDA.fhi, 28-Ni.LDA.fhi, 13-Al.GGA.fhi,
28-Ni.GGA.fhi.
What else could be wrong? Thanks for any help.
Best regards,
Stephen
This is one of my infiles: acell 5.7970482729504687 5.767560060000 5.767560060000 angstrom
ecut 480 eV
ionmov 0 ixc 1 kptopt 1 natom 16
ngkpt 10 10 10 nstep 15 ntypat 2 nband 65
occopt 4 tsmear 0.03
toldfe 0.005 eV
chkprim 0
znucl 13 28
typat 1
1
1
1
1
1
1
1
2
2
2
2
2
2
2
2
xangst 0.000000 0.000000 0.000000
0.000000 0.000000 2.883780
0.000000 2.883780 0.000000
0.000000 2.883780 2.883780
2.898524 0.000000 0.000000
2.898524 0.000000 2.883780
2.898524 2.883780 0.000000
2.898524 2.883780 2.883780
1.449262 1.441890 1.441890
1.449262 1.441890 4.325670
1.449262 4.325670 1.441890
1.449262 4.325670 4.325670
4.347786 1.441890 1.441890
4.347786 1.441890 4.325670
4.347786 4.325670 1.441890
4.347786 4.325670 4.325670
--
Chol-Jun Yu
Computational Materials Engineering (CME)
Institute of Minerals Engineering (GHI)
Center for Computational Engineering Science (CCES)
RWTH Aachen University (RWTH)
Jülich-Aachen Research Alliance (JARA)
Mauerstr. 5, D-52056 Aachen
Tel.: +49-241-8094989 Fax: +49-241-80695097
Attachment:
First principles calculations of elastic properties of metals.pdf
Description: Adobe PDF document
ndtset 11
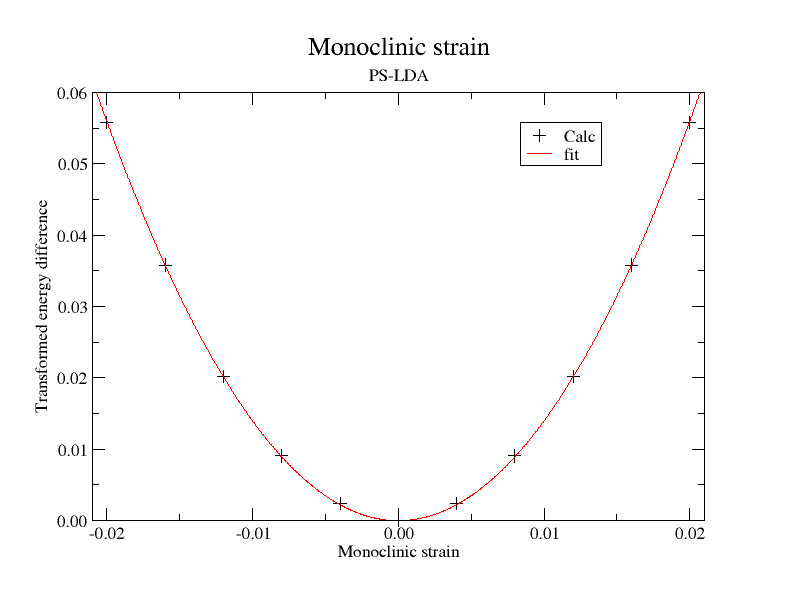
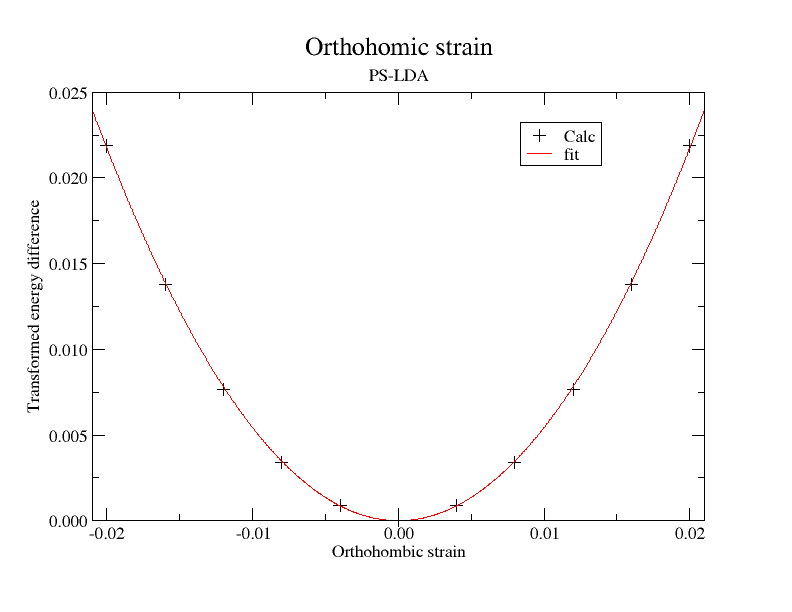
rprim1 1.0 -0.010 0.0 -0.010 1.0 0.0 0.0 0.0 1.000100010001
rprim2 1.0 -0.008 0.0 -0.008 1.0 0.0 0.0 0.0 1.000064004096262
rprim3 1.0 -0.006 0.0 -0.006 1.0 0.0 0.0 0.0 1.000036001296047
rprim4 1.0 -0.004 0.0 -0.004 1.0 0.0 0.0 0.0 1.000016000256004
rprim5 1.0 -0.002 0.0 -0.002 1.0 0.0 0.0 0.0 1.000004000016
rprim6 1.0 0.000 0.0 0.000 1.0 0.0 0.0 0.0 1.0
rprim7 1.0 0.002 0.0 0.002 1.0 0.0 0.0 0.0 1.000004000016
rprim8 1.0 0.004 0.0 0.004 1.0 0.0 0.0 0.0 1.000016000256004
rprim9 1.0 0.006 0.0 0.006 1.0 0.0 0.0 0.0 1.000036001296047
rprim10 1.0 0.008 0.0 0.008 1.0 0.0 0.0 0.0 1.000064004096262
rprim11 1.0 0.010 0.0 0.010 1.0 0.0 0.0 0.0 1.000100010001
acell 3*5.3516199522
ecut 54.0
occopt 4
tsmear 0.02
toldfe 1.0d-10
prtgeo 0
prtden 0
prteig 0
prtwf 0
kptopt 1
ngkpt 8 8 8
nshiftk 1
shiftk 0.0 0.0 0.0
nstep 30
ixc 7
ntypat 2
znucl 13 28
natom 2
typat 1 2
xred
0.0 0.0 0.0
0.5 0.5 0.5
ndtset 11
rprim1 0.980 0.0 0.0 0.0 1.020 0.0 0.0 0.0 1.000400160064026
rprim2 0.984 0.0 0.0 0.0 1.016 0.0 0.0 0.0 1.000256065552782
rprim3 0.988 0.0 0.0 0.0 1.012 0.0 0.0 0.0 1.000144020738986
rprim4 0.992 0.0 0.0 0.0 1.008 0.0 0.0 0.0 1.000064004096262
rprim5 0.996 0.0 0.0 0.0 1.004 0.0 0.0 0.0 1.000016000256004
rprim6 1.000 0.0 0.0 0.0 1.000 0.0 0.0 0.0 1.0
rprim7 1.004 0.0 0.0 0.0 0.996 0.0 0.0 0.0 1.000016000256004
rprim8 1.008 0.0 0.0 0.0 0.992 0.0 0.0 0.0 1.000064004096262
rprim9 1.012 0.0 0.0 0.0 0.988 0.0 0.0 0.0 1.000144020738986
rprim10 1.016 0.0 0.0 0.0 0.984 0.0 0.0 0.0 1.000256065552782
rprim11 1.020 0.0 0.0 0.0 0.980 0.0 0.0 0.0 1.000400160064026
acell 3*5.3516199522
ecut 54.0
occopt 4
tsmear 0.02
toldfe 1.0d-10
prtgeo 0
prtden 0
prteig 0
prtwf 0
kptopt 1
ngkpt 8 8 8
nshiftk 1
shiftk 0.0 0.0 0.0
nstep 30
ixc 7
ntypat 2
znucl 13 28
natom 2
typat 1 2
xred
0.0 0.0 0.0
0.5 0.5 0.5
- B2 NiAl, Stephen . Hocker, 02/20/2008
- Re: [abinit-forum] B2 NiAl, Chol-Jun Yu, 02/20/2008
- Re: [abinit-forum] B2 NiAl, Anglade Pierre-Matthieu, 02/20/2008
- Re: [abinit-forum] B2 NiAl, Chol-Jun Yu, 02/20/2008
- Re: [abinit-forum] B2 NiAl, Matthieu Verstraete, 02/20/2008
- Re: [abinit-forum] B2 NiAl, D. R. Hamann, 02/20/2008
Archive powered by MHonArc 2.6.16.